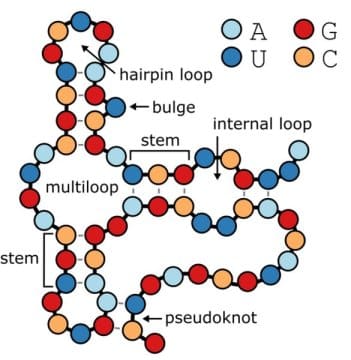
BpRNA software is a big data annotation tool for secondary structures in ribonucleic acids, which is capable of parsing RNA structures ending up with objective, precise, and easily interpretable descriptions of all loops, stems, and pseudoknots, providing positions, sequence, and flanking base pairs for each structural features enabling study of RNA structures en masse at large scale.
RNA works with DNA to produce proteins required throughout the body. While DNA contains hereditary information, RNA delivers informations coded instruction to protein manufacturing sites within cells. Noncoding RNAs do not encode protein. RNA is a fundamental essential molecule for life to exist, understanding their structure is important to understand their function.
Within single nucleic acid polymers or between two polymers secondary structures are base pairing interactions. DNA has fully based paired double helices, while RNA is a single strand and can form complicated interactions. According to researchers bpRNA features the largest detailed meta-database to date of over 100,000 complex secondary RNA structures, providing a colour coded map of where everything is. Annotations may enable identification of statistical trends which may shed new light on RNA structure formations opening new paths for machine learning algorithms to predict secondary RNA structures in ways not yet possible.
New RNAs are discovered frequently, much progress is being made in understand their functions, and gaining appreciation of the fact the genome consists of noncoding in addition to messaging RNAs which are important biological molecules each having big effects on human health and disease.